On July 9, the Crop Stress Tolerance and Genetic Improvement Team at the Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, in collaboration with domestic and international research institutions, constructed the first single-cell multi-omics atlas of multiple rice organs. The study systematically analyzed the functions of diverse cell types and their regulatory roles in complex traits, and predicted cell type–specific genes controlling important agronomic traits. These findings provide cell-level candidate targets for rice molecular design breeding. The related research was published in Nature.
Single-cell omics represents a major technological breakthrough in life sciences, offering unique advantages for revealing cellular heterogeneity and dissecting biological functions at an unprecedented resolution. Rice, one of China’s most important staple crops, is composed of various functional cell types in its roots, stems, leaves, and seeds. These cell types play essential roles in growth, yield, and stress resistance, yet their systematic identification and functional characterization have remained largely unexplored. By applying single-cell and spatial omics technologies, researchers can now characterize cell types and states across organs, and elucidate molecular regulatory mechanisms underlying rice development and key agronomic traits.
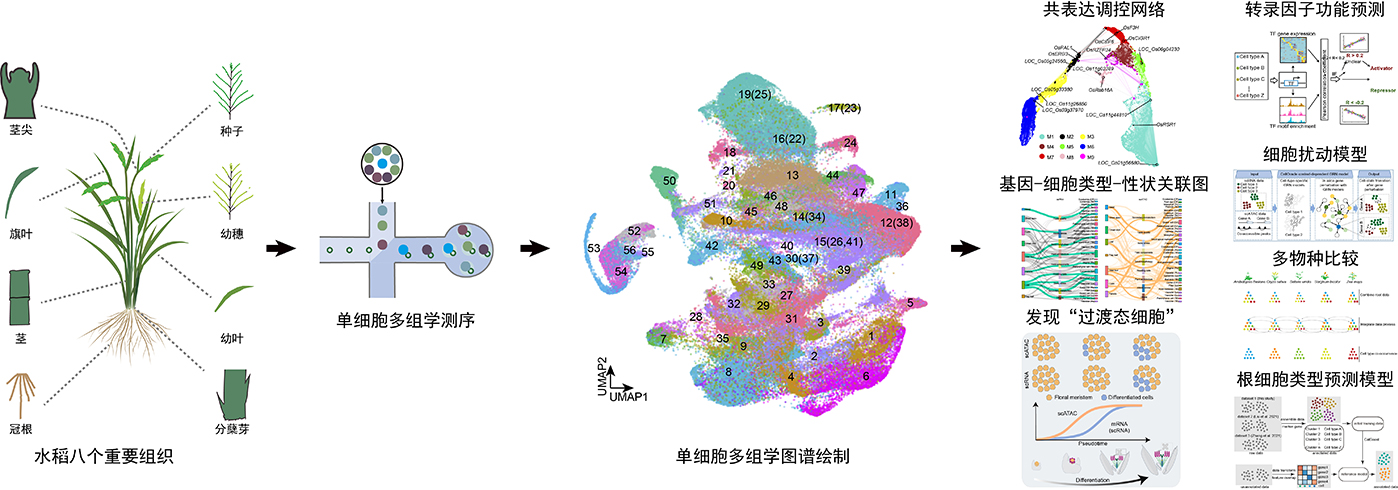
In this study, the researchers simultaneously profiled RNA expression and chromatin accessibility of over 110,000 cells from eight major rice organs. Through integration with in situ hybridization experiments, they identified 54 distinct cell types and constructed cell type–specific gene regulatory networks. The team discovered a previously unrecognized “transitional cell state” in the inflorescence meristem that plays a pivotal role in determining cell fate transitions. Using co-expression network analysis, motif enrichment prediction, and virtual gene knockout simulations, they identified and validated multiple key regulators functioning in specific cell types—for instance, RSR1 governing root cortex development, F3H coordinating carbon-nitrogen metabolism, and LTPL120 regulating tiller formation and plant architecture.
By integrating genome-wide association study (GWAS) data, the researchers further revealed correspondences between agronomic traits and specific cell types, and demonstrated both the conservation and diversification of cell functions during evolution. Additionally, the team released the Rice Single-Cell Multi-omics Resource (Rice-SCMR; http://www.elabcaas.cn/scmr), providing an open platform for functional gene discovery and intelligent breeding design.
This work establishes a technical foundation for intelligent design breeding at single-cell resolution, opening a new avenue for molecular crop improvement. It will strongly support national food security and the revitalization of the seed industry.
The research was supported by the National Key R&D Program of China for Biological Breeding, the National Natural Science Foundation of China, and the Central Public-interest Scientific Institution Basal Research Fund.
Original article: https://www.nature.com/articles/s41586-025-09251-0 |